How many clicks does it take to get to the center of an Electronic Health Record (EHR)? Some researchers have actually tried to find out. During a busy 10-hour shift, an emergency medicine physician will make 4,000 clicks to complete data entry tasks and chart review, leaving about 28% of their time for direct patient care.
If you’re like me, you might be wondering why it takes 6 clicks to order an aspirin (a first-line treatment for heart attack) or 47 clicks to document a physical exam for back pain (observed in 1 in 2 Americans). You might also be wondering how emergency medicine physicians tolerate such burdensome workflows every shift.
Generative artificial intelligence (AI) and specifically, large language models (LLMs), are transforming user experience in healthcare and life sciences. In a rather short period of time, LLMs have demonstrated striking competence with biomedical data retrieval and information synthesis. Conversational interfaces powered by LLMs are facilitating biomedical data access and information synthesis via natural language, catalyzing a new paradigm in biomedical human-computer interaction.
A paradigm I’m calling “Command Line Medicine.”
In this piece, I explore:
The origins of biomedical information architecture and design
Examples of how natural language can direct computational processes in biomedicine
Companies pioneering the transition to Command Line Medicine
Open questions and derivative predictions for how LLMs will inspire us to reimagine user experience in healthcare and life sciences
What is Command Line Medicine? A command line terminal is a text-based interface developed in the field of computer science that allows a user to interact directly with an operating system. Despite its simplicity, the command line grants users unparalleled control of core computing processes without abstraction.
The paradigm of Command Line Medicine will allow users of biomedical information systems to direct computational processes using natural language, thereby granting superpowers and efficiencies akin to those enjoyed by software developers. We will exchange 4,000-click complexity and clunky Graphical User Interfaces (GUIs) for conversational control of systems enabling robust data retrieval, analysis, and synthesis.
Due to the abundance of high-value structured and unstructured data sitting behind challenged user experiences, electronic health records (EHRs) have emerged as a compelling starting point for biomedicine’s transition to the command line.
Eventually, however, the AI-enabled command line will impact every major information system in healthcare and life sciences from laboratory information management to pharmacy to billing and coding to manufacturing execution, and more.
How does Command Line Medicine work in practice? Examples of natural language prompts for the EHR may take two shapes:
Requests for information (“read”):
“What is the updated troponin series for Patient X (MRN123456) over the last three days? What did the radiology report for this morning’s abdominal CT scan say?”
Instructions for the system to perform a specific function (“write”):
“Update MRN78901 progress note with the following dictation and create an order for aspirin 325mg stat.”
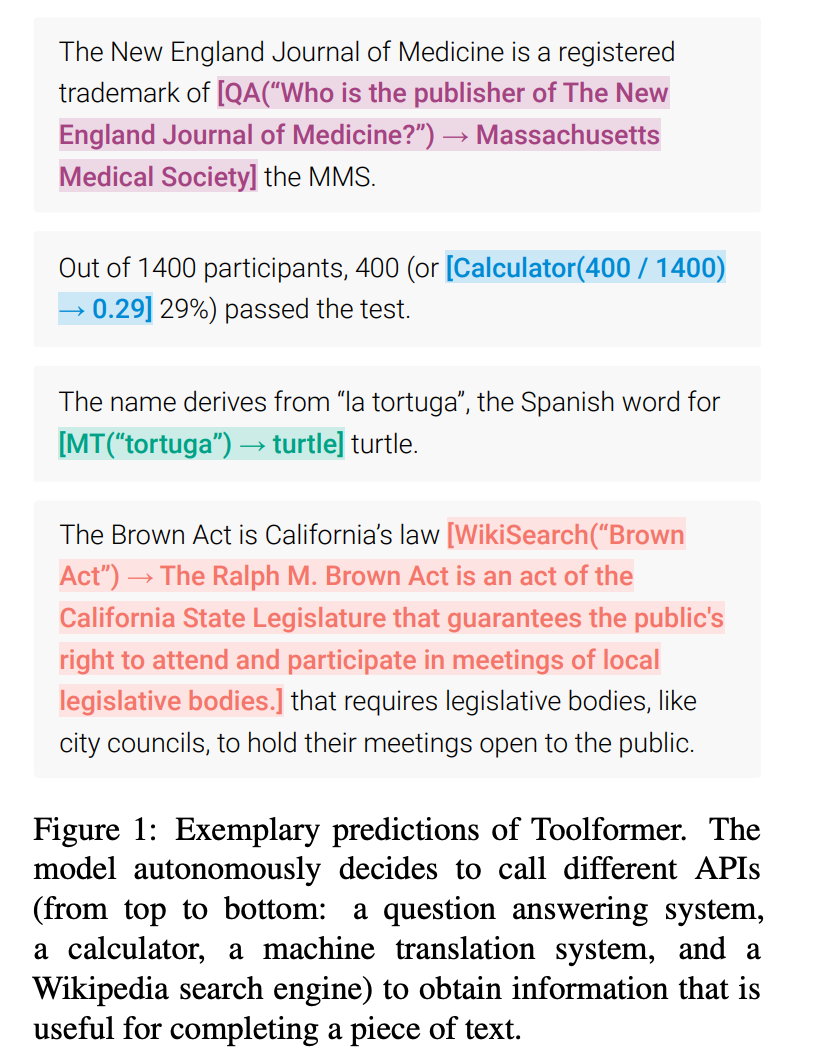
These directives are fairly straightforward use cases that are natural extensions of core LLM functionality, such as natural language querying and data labeling. Over time, integration of data from other systems will also become possible with Command Line Medicine. As we mature infrastructure for making API calls via LLMs — see ToolFormer, a model that ingests natural language instructions and decides which APIs to call, when to call them, and what arguments to pass — we will free ourselves from janky i-Frames superimposed over EHR GUIs in exchange for deeper embeddings of data in a unified view.
In the near future, users of biomedical information systems will access “App Store”-like functionality natively (see ChatGPT plugins). Instead of interfacing with dozens of browser tabs each requiring separate credentials — in the case of clinicians, that might include bouncing between platforms like UpToDate, MDCalc, PubMed, and the EHR — pre-trained language models accessed via API could offer access to a wide variety of data sources through a single interface, such as at the point of care through the EHR.
As an example of one “app” that might be accessible by the command line, consider being able to instantiate a medical calculation in the EHR using natural language:
How does this differ from today’s workflow? On MDCalc, calculating the Wells’ Score to assess the likelihood of pulmonary embolism requires up to 10 clicks and manual back-referencing with data dispersed throughout a patient’s chart. Should a clinician desire to copy the calculation into the EHR for documentation purposes, add two more clicks: one click to copy and another to paste. If you wanted to perform the calculation on UpToDate instead, you’d have to search for the articles on Pulmonary Embolism, locate the right one, and scroll until you find the clinical calculator. Compare this experience to that supported by ChatGPT using the clinical vignette above. These systems will also be capable of suggesting clinical calculations based on the vignette provided.
Note: some readers have flagged that the Wells’ Score is calculated incorrectly. I will caveat that this example is for demonstrative purposes, and by no means am I suggesting that ChatGPT is ready to be deployed in the clinic. Not missing the forest for the trees, it's conceivable that GPT plus smaller models fine-tuned for medical calculations (or even medical calculator APIs, accessible via ToolFormer) would be able to perform this task with high accuracy in the very near future.
Perhaps it is abundantly clear how clinicians surpass thousands of clicks per shift.
In the near future, LLMs will perform clinical calculation tasks with high fidelity and accuracy, automatically parsing key parameters from structured and unstructured notes to suggest and generate clinical scores with data provenance at the point of care.
As another example, consider how Command Line Medicine transforms medical information-seeking when compared to existing methods. On UpToDate, a question about myxedema coma will lead you to an article titled “Myxedema Coma.” However, it will not lead you to the answer to your question; instead, you must then scroll, search, and parse pages of text to find what you are looking for: epidemiology and risk factors, clinical presentation, diagnosis, treatment, monitoring, prognosis, and more.
In the era of LLMs, UpToDate has not yet implemented the capability to provide personalized responses to medical questions. Instead of landing on the Myxedema Coma page and navigating down to the dosing link, then navigating to levothyroxine vs. liothyronine, imagine being able to ask: “For patients with suspected myxedema coma, what are the initial and maintenance doses for levothyroxine and expected improvements in 48-72h T4?” right from the EHR. The system would output a tailored response (here is ChatGPT’s answer, without specific training for medical use cases):
Next, clinician users could immediately drop this information into their plans to drive downstream orders. In theory, LLMs could pre-populate orders directly from the above plan for review by the physician. This example demonstrates how LLMs could supercharge both diagnostic workflows and downstream plan-related tasks. Note: these applications require further research on AI performance and safety, and should be considered adjunctive in the interim. Together with researchers at the Stanford Institute for Human-Centered Artificial Intelligence, I’ve conducted research on the safety and utility of LLMs for meeting clinical information needs at the point of care - read more here.
The locus of control over biomedical information is shifting from inflexible graphical user interfaces toward natural language.
A Brief History of Command Line Medicine
How did we get here? To my surprise, Command Line Medicine is actually not a new concept. In many ways, we are returning to the first principles of biomedical information systems design first proposed in the 1970’s, when concerted efforts to interrogate how computers would transform medical data custodianship, retrieval, access, and physician experience blossomed. According to Cosby et al. in 1971, “the technique of physician-computer interaction gives the clinician or researcher enhanced diagnostic capabilities, unobtainable by batch processing or standard techniques of statistical analysis.”
After reviewing literature from this era, it’s clear that GUI-based medical technology was not always a dominant biomedical design paradigm, especially for EHRs. In the early days of computerized biomedical data management, technologists explored many information architectures including checklists, questionnaires, question-and-answer dialogues, and free narrative text recordings (documented by a human medical secretary).
One of the first published systems for computer-based authorship of progress notes allowed physicians to use a command line interface to enter and retrieve information. The Hypertension Clinic was the first site selected to test the system in 1970. To enter data, the physician would select a topic from a list of choices (e.g., chest problems), from which a series of additional options emerged (e.g., chest pain, arrhythmia, congestive heart failure, etc.). After selecting the right options from various levels within the tree, the physician could indicate normal/abnormal physical exam findings. This computerized system was skeuomorphic, in that it mirrored the ways clinicians are trained to think about clinical problems:
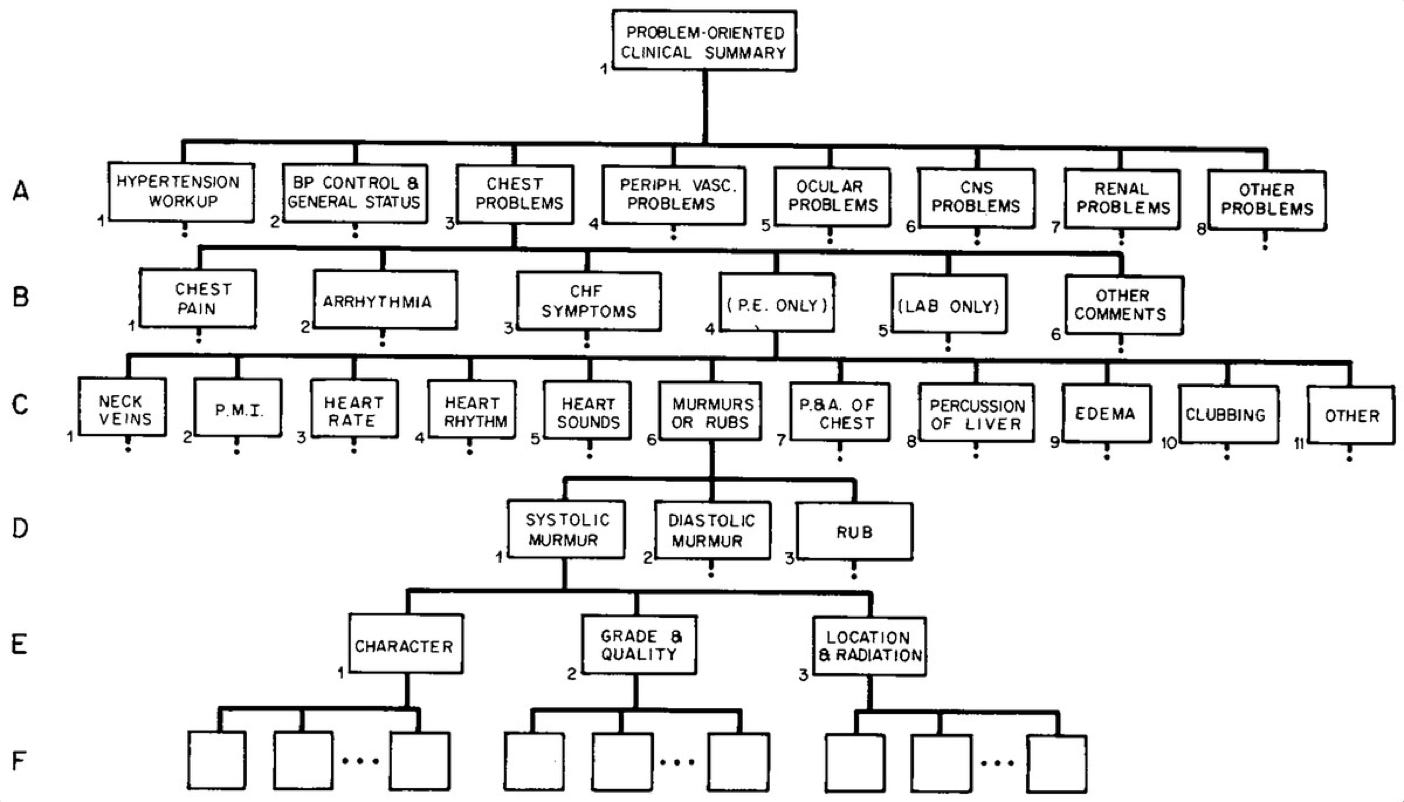
Upon completion, the progress note would be printed into the terminal to be signed by the physician, then inserted into the medical record with time-series context. To retrieve information, the physician could enter a statement describing something along the lines of “chest pain, frequency more than once a week, lasting more than five minutes and noted to be relieved by nitroglycerin.” In the retrieval mode, this request could locate all records meeting these criteria.
Impressively and despite the rigid rules-based architecture, this early instantiation of medical record technology demonstrated material workflow improvements, including a one-third reduction in time spent writing notes and an increased density of information to the tune of 2x more data compared to writing notes by hand. I’d be remiss not to mention that the system worked by cathode ray tube, such that the physician had to physically touch a metallic strip to “complete the circuit” and indicate their selection!
Medical research also pioneered command line interfaces for biomedical data retrieval. PaperChase, a computer program for searching medical literature, was first introduced in 1981. Back-ended by a medical library of 400,000 references, PaperChase satisfied information requests for clinicians and clinicians-in-training by allowing users to search medical literature by author's name, journal of publication, title word, or medical subject heading. Despite each search consuming an average of 78 seconds of processing, PaperChase demonstrated high user satisfaction across patient care, research, and teaching use cases.
Next is UpToDate, a leading medical reference tool founded in 1992. The platform has a rich and inspiring history dating back to its founder, an enterprising nephrologist named Dr. Bud Rose. You can read more about his story here. UpToDate is revered by some as “the most important medical invention in the past 30 years, possibly longer… and [Dr. Rose] should have won the Nobel Prize for medicine.”
UpToDate was one of the first medical resources to be adapted to a computerized format, despite Dr. Rose’s publisher declining to support the digitization of his flagship textbook, “Clinical Physiology of Acid-Base and Electrolyte Disorders.” Out of a desire to update his textbook more frequently than each new edition, Dr. Rose developed a version of his textbook on HyperCard and shared it with fellow physicians on floppy disks.
The most transformative feature of UpToDate was not technological in nature; it was how the articles were structured. UpToDate didn’t attempt to become a digitized encyclopedia of all human diseases. Rather, the platform proposed novel information architectures for medical content that optimized search efficiency and utility at the point of care. UpToDate’s content provides recommendations for clinician information-seekers pertaining to the diagnosis, treatment, dosing, monitoring, and more. You will not find a generic, Wikipedia-style article about Diabetes on UpToDate. But you will find articles that answer clinical questions about Diabetes diagnosis, treatment, and management. As discussed previously, while these answers are specific, they are not (yet) personalized based on a user’s question.
There is something beautiful about the simplicity and elegance of the biomedical information retrieval systems of the past. Of course, these systems were designed largely to handle uni-modal text data, whereas the systems of 2030 must support multi-modal data including text, imaging, video, and -omics.
The Next Generation of Command Line Medicine
As we return to the command line, several companies are pioneering biomedical human-computer interaction using LLMs:
Abridge
Abridge leverages command line infrastructure to automate documentation-related tasks for clinicians. After ambiently transcribing medical conversations between clinicians and patients using automated speech recognition, Abridge structures medical documentation for downstream “customers” of the data (e.g., consulting physicians, patients, billing specialists, etc.). The platform surfaces rich insights and metadata about the conversation in an editor tool, allowing clinicians to update the note, map key data elements to upstream conversational context, and identify opportunities to enrich the note based on the recipient.
Atropos Health
Atropos Health is an operating system for real-world evidence at the point of care. Physicians make hundreds of decisions per day. Some decisions are clearly defined by specialty and society guidelines, others exist in grey areas where guidelines are silent or where patients with a specific phenotype have not been studied. Leveraging large corpora of data stored in EHRs across the country, Atropos allows clinician users to query real-world data sets at the point of care using natural language in the form of PICO (Population / Problem, Intervention, Comparison, Outcome) questions. In the backend, Atropos can use code generation techniques enabled by LLMs to translate natural language queries into querying languages that interface efficiently with the EHR. The end result is a prognostogram that provides regulatory-grade evidence pertaining to the question. Imagine being able to produce a new, regulatory-grade real-world evidence study or clinical guideline in just a few sentences. This is the world Atropos is creating.
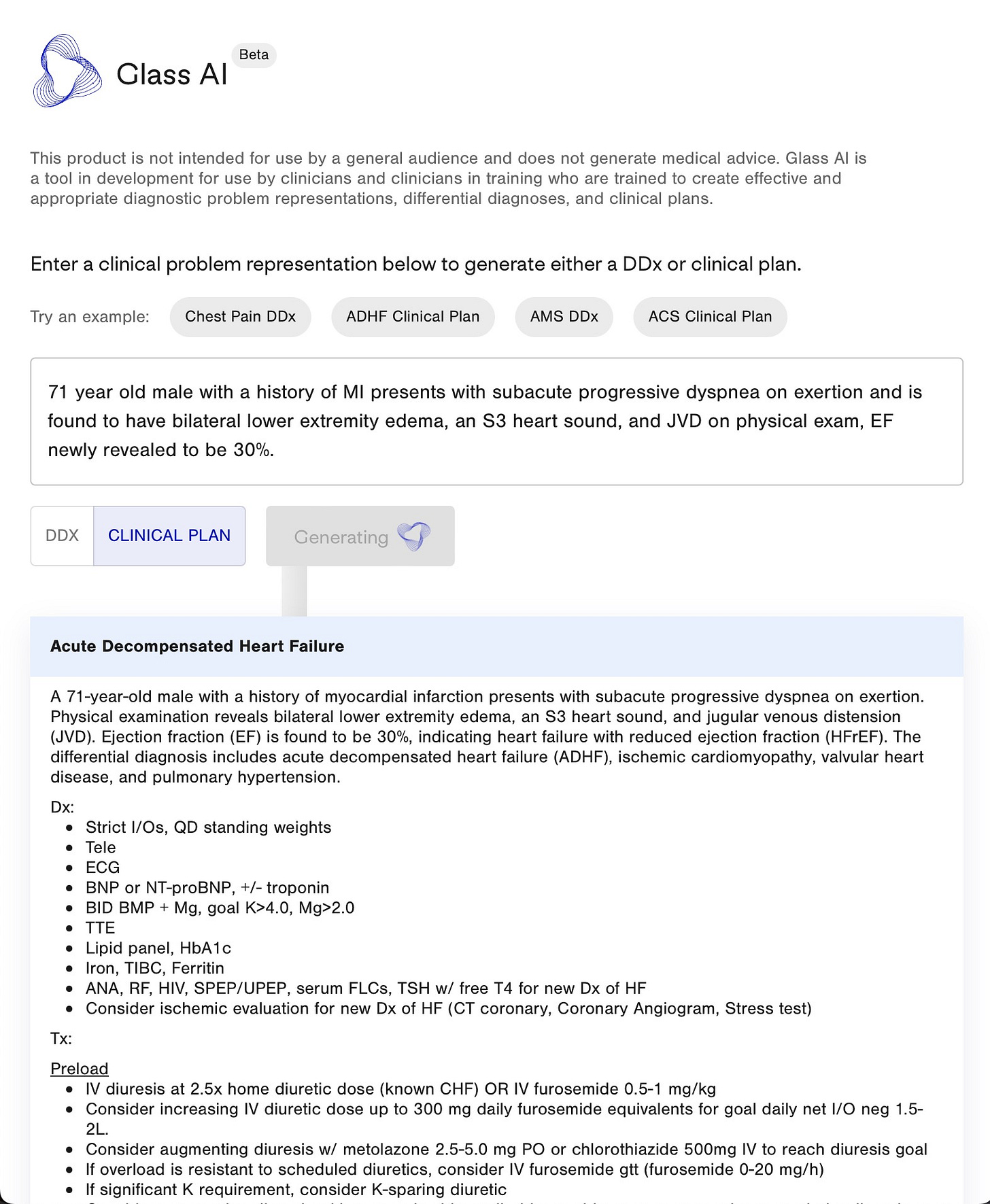
Glass Health
Glass Health is a “digital brain” for doctors that empowers clinicians to store, maintain, and connect medical knowledge in a single platform. Pro features of Glass include an AI-enabled differential diagnosis generator that ingests one-liner clinical problem representations and outputs full differentials and derivative clinical plans for each potential diagnosis.
Regard
Regard is a provider infrastructure and software company that integrates with EHRs to provide clinically actionable documentation adjustments that ensure complete and accurate capture of diagnoses in the medical record. The platform uses AI to surface relevant medical history from the patient chart into a user-friendly, command line tool that offers data provenance for any identified diagnoses.
Science.io
Science.io is a healthcare data infrastructure company that offers APIs for data structuring and redaction. Using Science.io’s tools, developers can create powerful natural language experiences with clinical data. As one example, Science created a command line Personal Health Record (PHR) whereby a patient could query their medical records using natural language.
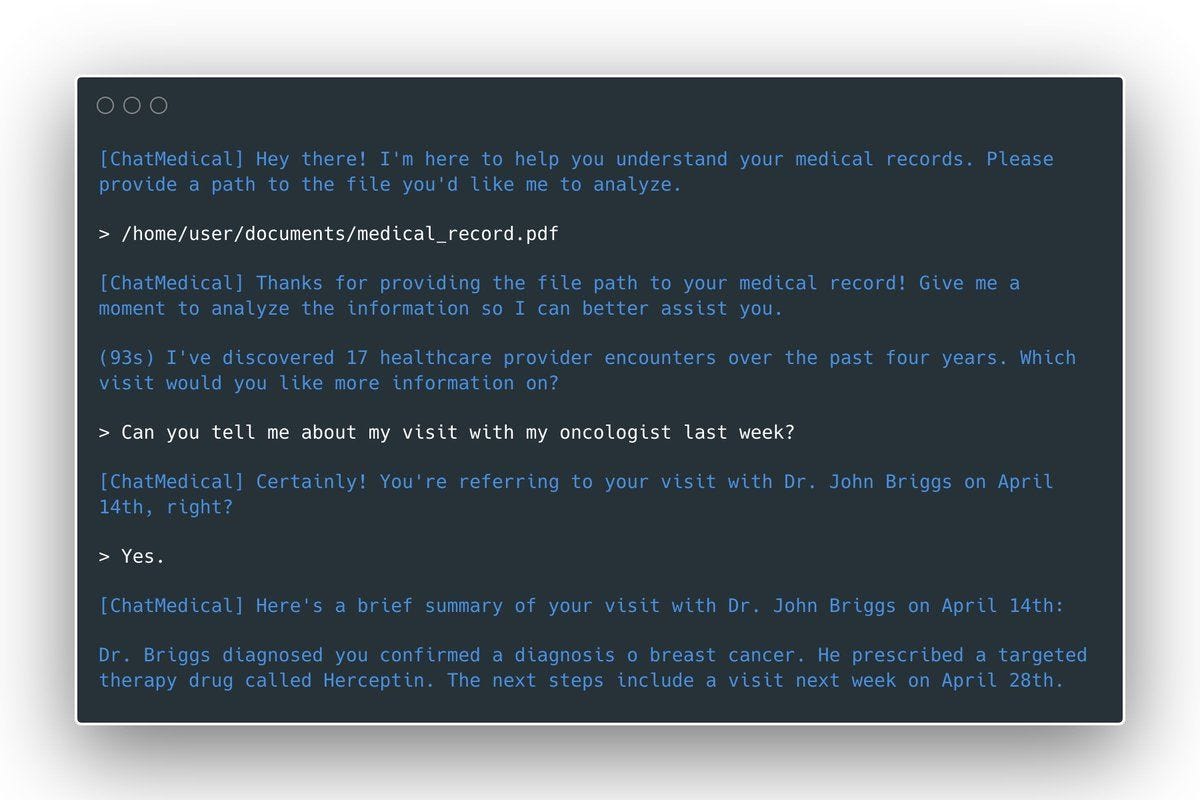
Epic
Epic is the largest EHR provider in the U.S. and also boasts a global presence with a client base that includes payers, pharma companies, and strategic partners. Cognizant of the ever-growing documentation burden shouldered by clinicians, Epic has made material investments to enhance productivity, launching features such as smart “Dot Phrases,” which allow commonly used chunks of text to be inserted easily into patient notes or discharge instructions by typing a period (the dot “.”) followed by a short phrase. For example, “.inpatientlabs” is a dot phrase that could pull relevant labs for a patient’s current inpatient hospitalization into a note in a tabular format.
In April 2023, Epic and Microsoft announced a partnership to explore applications of generative AI in electronic health records. Researchers at Stanford, UC San Diego Health, and UW Health became among the first to pilot these new tools with initial use cases focused on automating asynchronous communication in Epic InBasket, a messaging portal between clinicians and patients. By auto-populating responses to basic questions such as refill requests for review by a human clinician, this collaboration seeks to reduce the correspondence burden for clinicians.
Future research should investigate safety and bias, response quality and accuracy, time savings, and clinician and patient satisfaction with LLM auto-drafting. A recent study by Ayers et al. in April 2023 found that in response to patient questions, ChatGPT produced higher quality responses 79% of the time compared to 22% for human physicians (a 4-fold difference), with empathy scores for ChatGPT responses 10x greater than human physicians.
Reimagining EHR search is another compelling opportunity for Command Line Medicine. Data duplication is an immense shortcoming of many EHRs, including Epic, where researchers have demonstrated that more than half of the total text in a given patient’s record is duplicated from prior text written about the same patient. These results stem from a well-intentioned “copy forward” feature that allows clinicians to copy a previous note for a patient into a separate one.
Unfortunately, this feature has had unintended consequences that have resulted in information overload, making it more difficult to find relevant information about a patient quickly. These challenges are further exacerbated by the click-heavy GUIs of most EHRs. In response, I have heard stories of doctors setting up VPNs and paying nurses in countries outside of the U.S. to pre-round on their patients, informing them of real-time chart updates by text.
LLMs will enable EHR users (e.g., clinicians, administrators, and back-office specialists) to query clinical data at scale using natural language. This transition will mark one of the greatest transformations of human-computer interaction in medicine to date.
Challenges with Command Line Medicine
Though there is much to be excited about in freeing medical information from the tyranny of click-based interfaces and outdated GUIs using LLMs and other generative models, Command Line Medicine poses new risks. Below I outline some of the questions I’m most invested in seeing the field answer over the coming years.
How do we teach models to adhere to human conventions that ensure patient safety, such as “closed-loop communication”? For example, an attending physician asks a resident to put in an order for a specific medication during a code. Using closed-loop language, the resident would verbally restate the medication, dosage, and delivery mechanism before administering it to ensure sure they have understood the order correctly. What does closed-loop communication look like between a model and a human doctor? Integrating and adapting conventional safety protocols, in addition to identifying new AI-native safety protocols, will be essential for LLMs to be effective in clinical environments.
What are the malpractice risks and/or benefits of utilizing LLMs for clinical information retrieval and decision-making? Interestingly, ChatGPT has been blocked by some hospitals already.
How do we evaluate the performance of foundation models such as LLMs in medicine? Though ChatGPT passing the US Medical Licensing Exam (USMLE) is striking, the USMLE is not the right evaluation task for understanding model performance on specific clinical tasks. In fact, many have argued it’s not the right evaluation criteria for human physicians, but I digress! The field needs to develop task-specific benchmarks for performance across use cases such as clinical decision support, billing and coding, clinical information retrieval, and more.
Should we give patients command line access to biomedical information systems? The user experience benefits of the biomedical command line will accrue to both clinician and patient users, and in many ways, align with recent Information Blocking rules. LLMs are uniquely suited to de-jargon medical information for patients by summarizing information at varying literacy levels. But what happens when an LLM provides misinformation? What is the medicolegal chain of responsibility?
Can we navigate the balancing act between summarization and data preservation? The aforementioned use case relies on summarization – a powerful capability of LLMs that utilizes a technique called compression, which is the process of condensing information that often involves rewriting sentences, combining ideas, or omitting details. In many settings, compression can abstract complexity for ease of understanding for a human reader. In a medical context, however, these techniques could result in data loss that has deleterious effects. For example, a patient asking questions about their medical record may not need to know exact lab values, but a clinician does. Ensuring biomedical factual consistency with summarization tasks will be key to ensuring safety.
How will we measure and address issues of hallucination in real-world clinical settings? Sometimes LLMs produce content that is incorrect or non-sensical, which is referred to as “hallucination.” In truth, hallucination is more of a feature than a bug of LLMs, and there are many methods for mitigating it outlined succinctly in this article.
How can we ensure privacy preservation with Protected Health Information as LLMs become connected to more databases and other models? Researchers at Deepmind rightfully called into question the risks of using LLMs trained on sensitive data, such as PHI. Even with appropriate redaction techniques, concerns loom that savvy prompt engineering could allow a user to back into private or safety-critical information used to train the model with relative ease, potentially allowing for the re-identification of patients by predicting underlying sensitive data (e.g., genomic, clinical, etc.).
How should foundation models be regulated? Foundation models by definition are able to perform a wide array of tasks. How do we ensure regulatory compliance for relevant tasks without holding back the progress of others? The FDA’s guidance on what constitutes a clinical decision support tool is hazy at best.
What are the human factors and ergonomics of AI in medicine? What derivative systems and processes require re-architecting as we transition back to the command line?
Predictions
The above risks and unanswered questions are not intractable problems. I expect that we will have much greater clarity in the next few years. Biomedicine’s transition to the command line is a matter of when and not if.
Here are a few of my predictions:
The field of medicine will run the largest Reinforcement Learning with (Expert) Human Feedback RL(E)HF experiment. We will learn more about best practices for integrating expert feedback into foundation models such as LLMs, and will benefit from investment in more robust infrastructure for doing so. I’ll be writing more on this in a separate piece.
The surface area for biomedical data capture explodes and becomes irrelevant. Data generation in biomedicine has become increasingly multi-faceted, a far cry from the 1970’s medical secretary-centered model. How data is captured will become less relevant over time as LLMs become increasingly adept with biomedical data structuring and normalization. Whether a patient encounter is captured by audio or a human scribe, LLMs will underpin the normalization of biomedical data across systems to support unified access via the command line.
Human-computer interaction will emerge as one of the most important areas of medical research. Fields such as pathology and radiology will shine a light on how the increasing augmentation of clinicians via technology and AI transforms the scope of clinical roles at a fundamental level, potentially creating entirely new roles while eliminating others.
Biomedical data structures and graphical user interfaces will unbundle and support enduring efforts for driving interoperability. Historically, data types and representations have been linked in healthcare. A simple example is viewing images. Oftentimes, users seeking access to a radiographic image are required to leave the EHR and enter the PACS system, a transition that has become increasingly smooth over time but still remains clunky. Our ability to manipulate data based on the information seeker’s needs and queries will create one-to-many relationships between a data type and its many representations. Ultimately, power will shift away from vertical platforms that have differentiated on a single interface (think data dashboarding companies), and will instead accrue to platforms with distribution that can harmonize data across sources and expose information via natural language formats.
Small models will remain key for task-specific optimization. Large models working collaboratively with small models > large models working alone. As we define “jobs to be done” by LLMs with increasing clarity and articulate benchmarks for performance, smaller task-specific models trained on proprietary data sets will ride shotgun with foundation models and provide material performance benefits.
Clinicians will be trained in biomedical prompt engineering, and yes, there will be prompt engineering Continuing Medical Education (CMEs).
Companies with existing distribution will enjoy a material headstart. I’m a big believer that the best products don’t win in healthcare and life sciences, but the best distribution does. Lifetime values for healthcare and life sciences software companies are notoriously long, as the operational costs of adding new vendors are high for industry incumbents. One of the greatest competitors that new healthcare technology companies face is actually incumbent systems, regardless of whether they are operating in the same category.
It is often easier for health system executives to wait 18-24 months for Epic to launch a capability than to onboard a new startup. Irrespective of what problem you are solving for a provider, all software tools are competing for the same budget at margin-scarce customers. So, while I am excited about emerging companies deploying LLMs across healthcare and life sciences, in the nearest term, I’m also extremely bullish on incumbents with existing distribution, unique data sets not-yet-seen by LLMs, and teams of specially trained workforces uniquely well-positioned RL(E)HF.
Product-led growth (PLG) models will flourish. Despite the distribution, data, and workforce advantages enjoyed by incumbents, there is immense opportunity for emerging healthcare and life sciences. As I’ve talked about in the past, companies that achieve rapid scale in healthcare and life sciences over the next decade may not be sold like those of the last. ChatGPT was the first machine-learning model to go viral, eclipsing 100 million users within two months post-release. Though GPT had been developed years prior to ChatGPT’s launch, the magic of the chatbot user experience made the underlying model accessible and inspired users to engage. As clinicians, scientists, and other healthcare and life sciences professionals seek solutions to their problems, I expect similar patterns to emerge, and companies reaching prospective users via PLG motions will thrive.
If you’ve made it this far, perhaps the most important prediction of them all comes from Dr. John Halamka, President of the Mayo Clinic Data Platform:
[re: AI and LLMs] “This technology won't replace doctors. But doctors who use AI will probably replace doctors who don't."
Swap “doctors” for any other profession in healthcare and life sciences and I believe the statement holds.
Conclusion
About 50 years ago, we began accessing biomedical information systems via command line interfaces. In 2023, we make a full return. As AI transforms the ways we access biomedical data, synthesize information, and make clinical and scientific decisions, many foundational principles will remain true. I’ll close by sharing those penned by Greenes et al. in their hallmark 1970 paper, “Recording, Retrieval and Review of Medical Data by Physician-Computer Interaction”:
The long-term effect of this technology on the character of medicine is difficult to estimate. Dramatic upheavals such as the abandonment of existing institutions and creation of new medical-care "utopias" are unlikely. Medical care will instead evolve gradually under the influence of current efforts. It is our conviction that systems such as that described in this paper have become necessary for medical-record management, for patient care and for research. It is imperative that models be developed that can permit operational evaluation and experience with some of the innovative and hitherto untested approaches to the problems of medical-record management that technology can make possible. We are no longer in a phase of demonstration and expostulation about what computers can do.
Thanks for reading,
Morgan
Thanks to my pals Chase Feiger, Brigham Hyde, and Nikhil Krishnan for their feedback on this piece.
Disclosures: I am an investor in Abridge.




















I very much enjoyed the piece. However, I don't think there will be a choice between command line interface and GUI interface. The future will be hybrid, highlighting the strengths of both interfaces.